McCrary Sorting Test
dc_test.Rddc_test implements the McCrary (2008) sorting test to identify violations of assignment rules.
It is based on the DCdensity function in the "rdd" package.
Arguments
- runvar
A numeric vector containing the running variable.
- cutpoint
A numeric value containing the cutpoint at which assignment to the treatment is determined. The default is 0.
- bin
A numeric value containing the binwidth. The default is
2*sd(runvar)*length(runvar)^(-.5).- bw
A numeric value containing bandwidth to use. If no bandwidth is supplied, the default uses bandwidth selection calculation from McCrary (2008).
- verbose
A logical value indicating whether to print diagnostic information to the terminal. The default is
TRUE.- plot
A logical value indicating whether to plot the histogram and density estimations The default is
TRUE. The user may wrap this function in additional graphical options to modify the plot.- ext.out
A logical value indicating whether to return extended output. The default is
FALSE. WhenFALSEdc_testwill return only the p-value of the test, but will print more information. WhenTRUE,dc_testwill return and print the additional information documented below.- htest
A logical value indicating whether to return an
"htest"object compatible with base R's hypothesis test output. The default isFALSE.- level
A numerical value between 0 and 1 specifying the confidence level for confidence intervals. The default is 0.95.
- digits
A non-negative integer specifying the number of digits to display in all output. The default is
max(3, getOption("digits") - 3).- timeout
A non-negative numerical value specifying the maximum number of seconds that expressions in the function are allowed to run. The default is 30. Specify
Infto run all expressions to completion.
Value
If ext.out is FALSE, dc_test returns a numeric value specifying the p-value of the McCrary (2008) sorting test.
Additional output is enabled when ext.out is TRUE.
In this case, dc_test returns a list with the following elements:
- theta
The estimated log difference in heights of the density curve at the cutpoint.
- se
The standard error of
theta.- z
The z statistic of the test.
- p
The p-value of the test. A p-value below the significance threshold indicates that the user can reject the null hypothesis of no sorting.
- binsize
The calculated size of bins for the test.
- bw
The calculated bandwidth for the test.
- cutpoint
The cutpoint used.
- data
A dataframe for the binning of the histogram. Columns are
cellmp(the midpoints of each cell) andcellval(the normalized height of each cell).
References
McCrary, J. (2008). Manipulation of the running variable in the regression discontinuity design: A density test. Journal of Econometrics, 142(2), 698-714. doi:10.1016/j.jeconom.2007.05.005 .
Drew Dimmery (2016). rdd: Regression Discontinuity Estimation. R package version 0.57. https://CRAN.R-project.org/package=rdd
Examples
set.seed(12345)
# No discontinuity
x <- runif(1000, -1, 1)
dc_test(x, 0)
#> Binwidth:
#> 0.03597
#>
#> Bandwidth:
#> 0.5025
#>
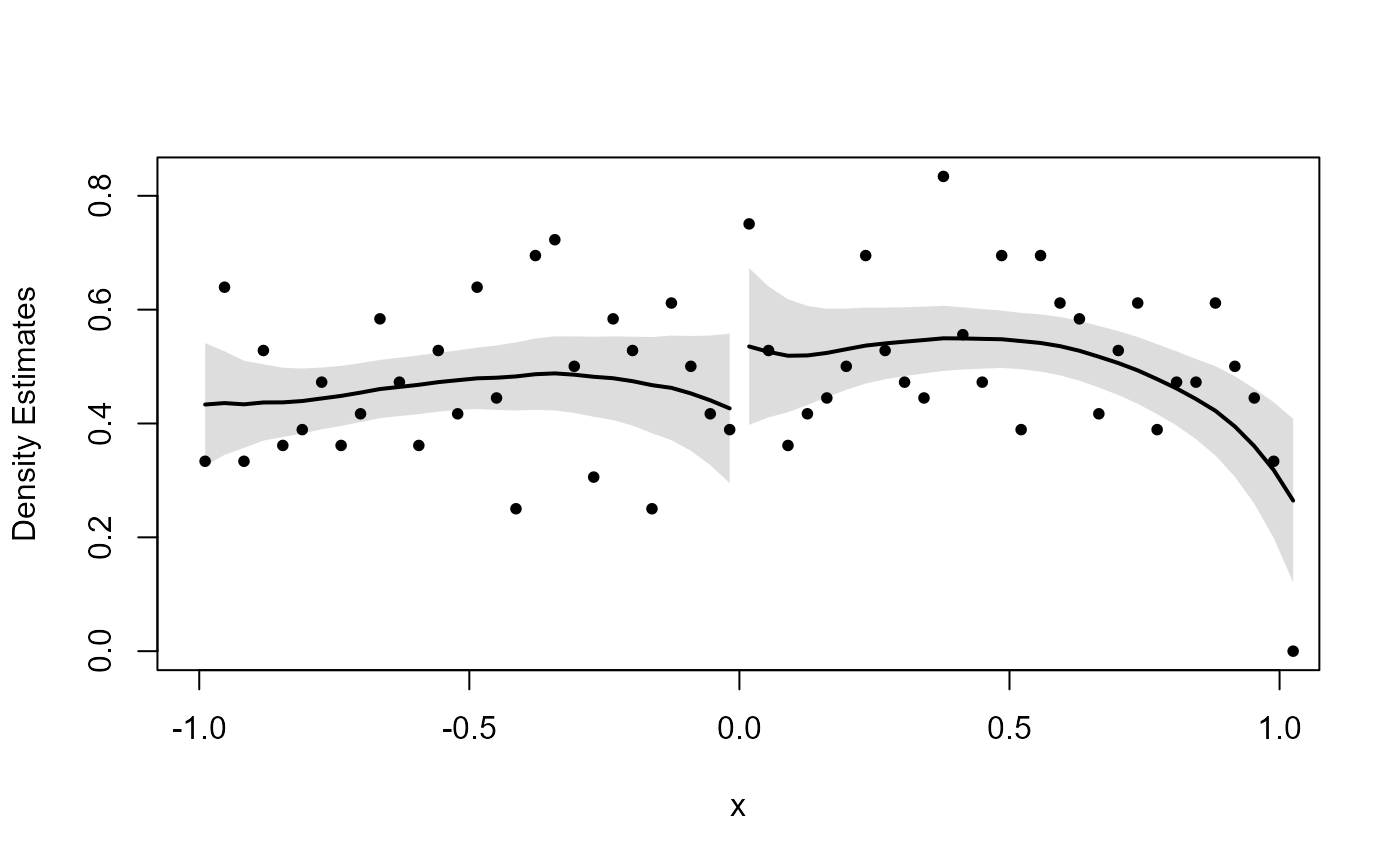 #> Estimate for log difference in heights:
#> Estimate Std. Error lower.CL upper.CL z value Pr(>|z|)
#> 0.2472 0.2011 -0.1469 0.6413 1.2294 0.2189
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Confidence interval used: 0.95
#>
#> [1] 0.2189345
# Discontinuity
x <- runif(1000, -1, 1)
x <- x + 2 * (runif(1000, -1, 1) > 0 & x < 0)
dc_test(x, 0)
#> Binwidth:
#> 0.04767
#>
#> Bandwidth:
#> 0.6016
#>
#> Estimate for log difference in heights:
#> Estimate Std. Error lower.CL upper.CL z value Pr(>|z|)
#> 0.2472 0.2011 -0.1469 0.6413 1.2294 0.2189
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Confidence interval used: 0.95
#>
#> [1] 0.2189345
# Discontinuity
x <- runif(1000, -1, 1)
x <- x + 2 * (runif(1000, -1, 1) > 0 & x < 0)
dc_test(x, 0)
#> Binwidth:
#> 0.04767
#>
#> Bandwidth:
#> 0.6016
#>
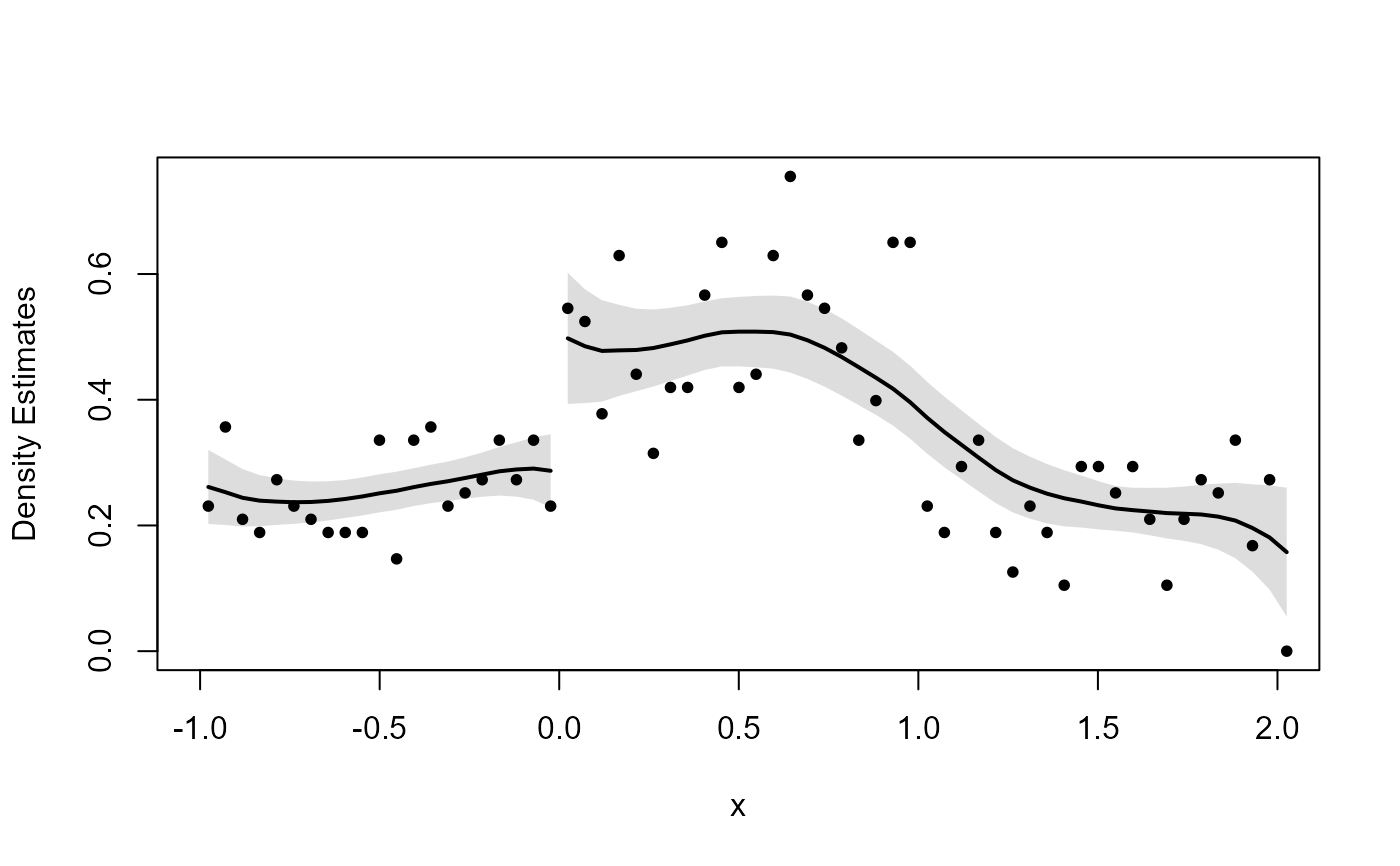 #> Estimate for log difference in heights:
#> Estimate Std. Error lower.CL upper.CL z value Pr(>|z|)
#> 0.56818 0.20925 0.15806 0.97830 2.71536 0.00662 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Confidence interval used: 0.95
#>
#> [1] 0.006620445
#> Estimate for log difference in heights:
#> Estimate Std. Error lower.CL upper.CL z value Pr(>|z|)
#> 0.56818 0.20925 0.15806 0.97830 2.71536 0.00662 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Confidence interval used: 0.95
#>
#> [1] 0.006620445